Definition
A stop codon is a genetic code that signals the end of protein manufacturing inside the cell, like a period at the end of a sentence. The three stop codons are nucleotide base triplets that play an important role in intracellular protein synthesis; physiological and/or anatomical changes are possible if a stop codon is in the wrong position on a DNA or RNA strand, or if the code sequence is changed.

What is a Stop Codon?
A stop codon is a single nucleotide triplet that provides an end-point for protein synthesis. If you already know something about how proteins are made inside the cell (do ribosomes and transfer RNA ring a bell?), you can imagine the line of messenger RNA working its way through a ribosome. A stop codon tells the ribosome and transfer DNA that the process can stop and the new polypeptide chain can be released. If ribosomes and transfer DNA are still a mystery, either read on or visit the protein synthesis page.
Without stop codons, an organism is unable to produce specific proteins. The new polypeptide (protein) chain will just grow and grow until the cell bursts or there are no more available amino acids to add to it. Both start and stop codons in DNA and RNA, just as their names suggest, provide start and stop instructions that regulate the length of a polypeptide chain. Each chain is the result of single amino acids connected in a particular order, as seen below.
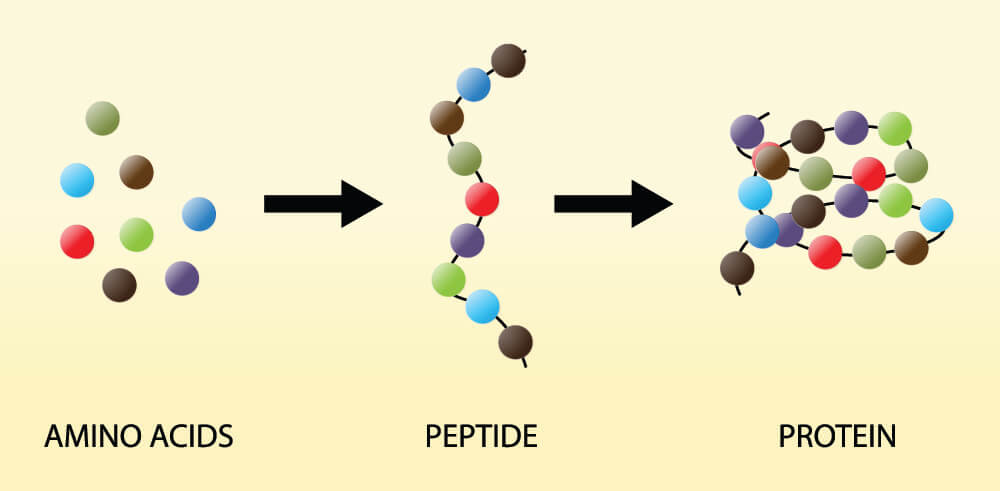
Genes are chemical codes that give groups of cells the instructions for protein production (coding genes) or decide when to make these protein-producing codes available (non-coding genes). When we think of the word codon we should immediately think of protein synthesis and our DNA. All organisms have a lot of non-coding (non-protein producing) information in their DNA but this subject needs its own article. When learning about the stop codon, it is enough to know that a codon is a three-letter code. This code either matches an amino acid or tells the cell when it is time to start (start codon) or stop (stop codon) adding amino acids to a polypeptide chain.
All codons are made up of three nucleotide bases and named according to the order of these bases – for example, the stop codon TAG tells us that it is made up of thymine, followed by adenine, followed by guanine. To truly understand the importance of the stop codon it’s useful to refresh our knowledge of DNA construction and protein synthesis.
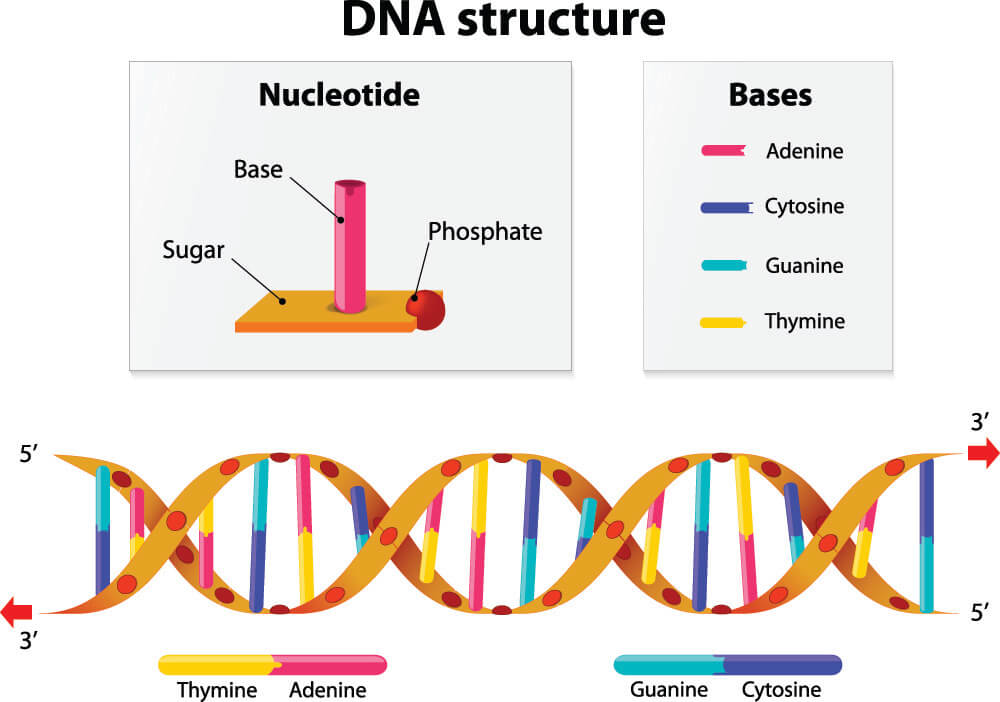
Stop Codons in Protein Synthesis
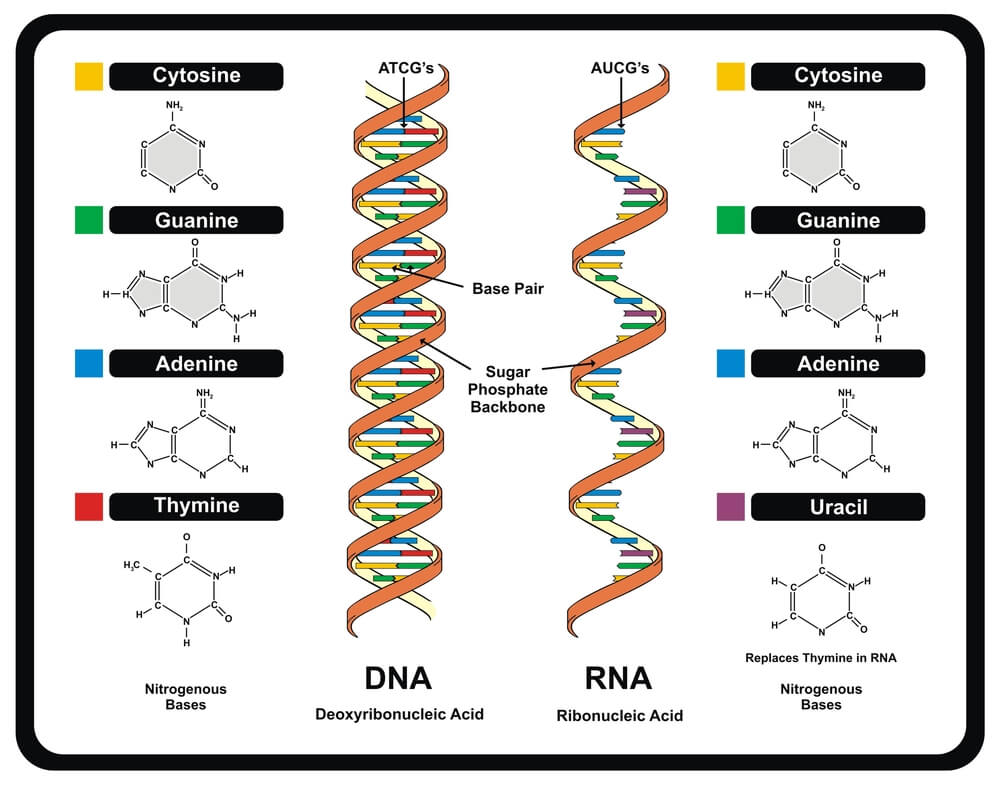
In protein synthesis, stop codons play an essential role. Every organism’s DNA is constructed from sugar, phosphate, and the nucleotide bases cytosine (C), guanine (G), adenine (A), and thymine (T). DNA forms a double-helix structure of two connected strands; the nucleotide bases provide the connection points, each attaching to specific partners. Two identical bases cannot connect, such as thymine to thymine or adenine to adenine. These bases are also extremely limited as to their choice of partner. Thymine attaches to adenine, adenine to thymine, guanine bonds with cytosine, and cytosine with guanine. It is important to remember these fixed partners: TA/AT and CG/GC. Things get a little more complicated when portions of DNA code are copied onto a strand of RNA during the transcription process of protein synthesis. When this happens, uracil replaces thymine; the fixed partners in RNA are UA/AU and CG/GC.
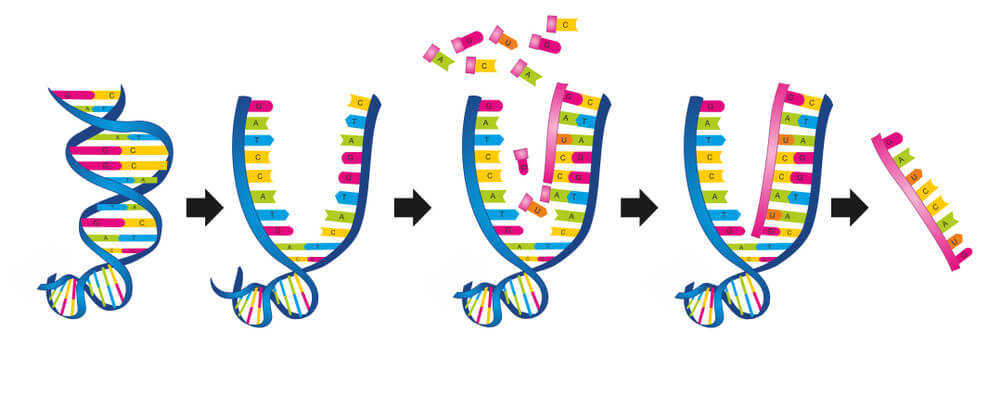
Stop Codons and Transcription
Gene sequences often run up to thousands of nucleotides in length. This means that figuring out the genetic code of any organism can be a real headache for scientists. To make things easier, when they identify a gene or allele they give it the equivalent of a GPS code – a cytogenic location. For example, the gene that shows a cell how to manufacture type I keratin 9, a protein found in the skin of our hand palms and foot soles, is found at cytogenic location 17q21.2.
The cytogenic location tells scientists where to locate different protein-manufacturing instructions. It is also important to remember that, while every cell nucleus contains the instructions to produce a fully-functioning organism, most genes are only expressed (activated) in specific tissues; KRT-9 is expressed in the skin cells of the palms and soles, while a liver cell nucleus also contains the instructions for keratin 9 production but the gene is not expressed.

Only around 1% of the human genome codes for proteins; it is thought that the rest of the code regulates how and when these proteins are produced. Non-coding DNA – previously called junk DNA – is now understood to monitor and control protein synthesis. It is noncoding DNA in combination with certain protein factors that decides when it is time for the double-helix structure to unzip so that the recipe (KRT-9) for type I keratin 9 can be copied, taken out of the nucleus, and sent to a ribosome so that this protein can be produced. Once the DNA has unzipped, a single strand of pre-messenger RNA is produced, transcription factors dictate when the copying process ends, the DNA zips back up and the now-complete mRNA strand moves out of the nucleus.
Although lots of sources talk about a mirrored mRNA copy, they do not always mention that this is a mirrored copy of a mirrored copy and, therefore, an exact copy of the coding strand of DNA. This is easier to understand when you consider that DNA is constructed from two separate strands – the coding (sense) strand that runs one way and the template (antisense) strand that runs antiparallel to it. If, for example, the sense strand runs left to right, the antisense strand runs right to left. If the sense strand contains the sequence AAAGCC, the antisense strand will consist of partner nucleotides running in the opposite direction: GGCTTT. RNA then transcribes (copies) the code of the antisense strand in an antiparallel fashion, so from left to right – exactly like the sense strand. That means the code of the RNA will be AAAGCC – exactly the same as the code of the antisense strand of DNA. There is only one potential difference – the partner of adenine in DNA is thymine but in RNA, thymine is replaced with uracil.
Once these attachments have been made during the process of DNA transcription, the RNA strand is renamed messenger RNA or mRNA.
A DNA start codon usually carries the code ATG (in mRNA, this is AUG), although other codons have been discovered that also initiate the translation of genes such as GUG and UUG. The code AUG also produced an amino acid called methionine so other factors in the form of enzymes play a role in knowing when a start codon is, in fact, a start codon and not just another amino acid to add to the polypeptide chain. Messenger RNA does not understand the meaning of the start codon, whatever code this happens to be. It simply copies it. The start codon is only of use during the translation phase of protein synthesis and the same applies to the stop codon.
Scientists agree that there are three stop codons – also called nonsense codons or termination codons – in the human genetic code. These are TAG, TAA, and TGA (DNA) and UAG, UAA, and UGA (RNA). Again, TAG, TAA, and TGA do not act as stop codons during transcription but are copied (substituting thymine for uracil) by RNA. Stop codons neither code for an amino acid nor belong to the non-coding group of genes but are a separate entity. Their recognition is much simpler than the recognition of the start codon. While the start codon also codes for an amino acid called methionine, stop codon amino acids do not exist; their triplet nucleotide sequences do not encode part of a polypeptide chain but only act to end the transcription and translation processes.
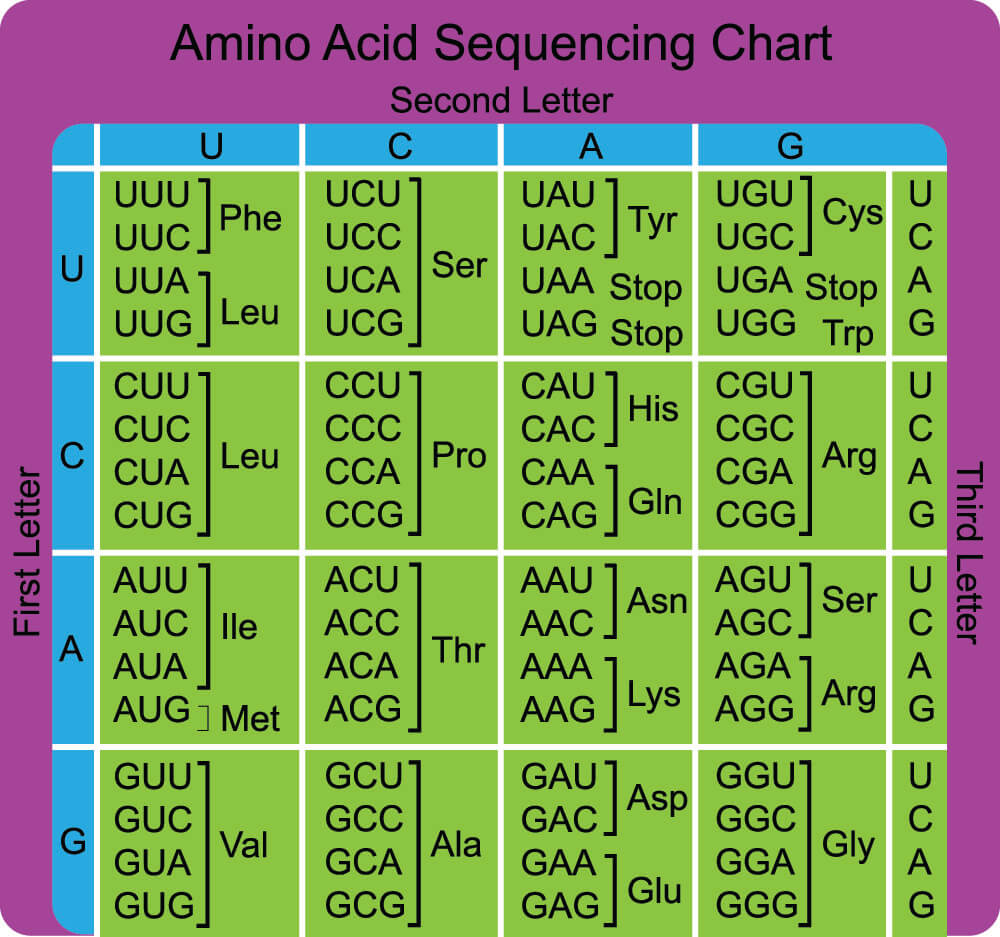
Stop Codons and Translation
The mRNA strand leaves the nucleus of the cell and travels to the cytoplasm, endoplasmic reticulum, or mitochondria where it enters between the subunits of a ribosome. The first codon ‘docking station’ inside a ribosome is the correct fit for methionine or one of the other nucleotide triplets that can play the part of start codon as well as code for an amino acid. This famous paper is two decades old, but we still don’t have a complete list of non-methionine start codons.
Once the start codon of the mRNA has been detected it is time for transfer RNA to bring the right amino acids in the same order as their associated nucleotide triplets. Each tRNA carries an amino acid that corresponds with a codon on the mRNA. Transfer RNA or tRNA ‘reads’ the codons of mRNA, which is why this step of protein synthesis is called translation. It is during the translation phase that start and stop codons function.
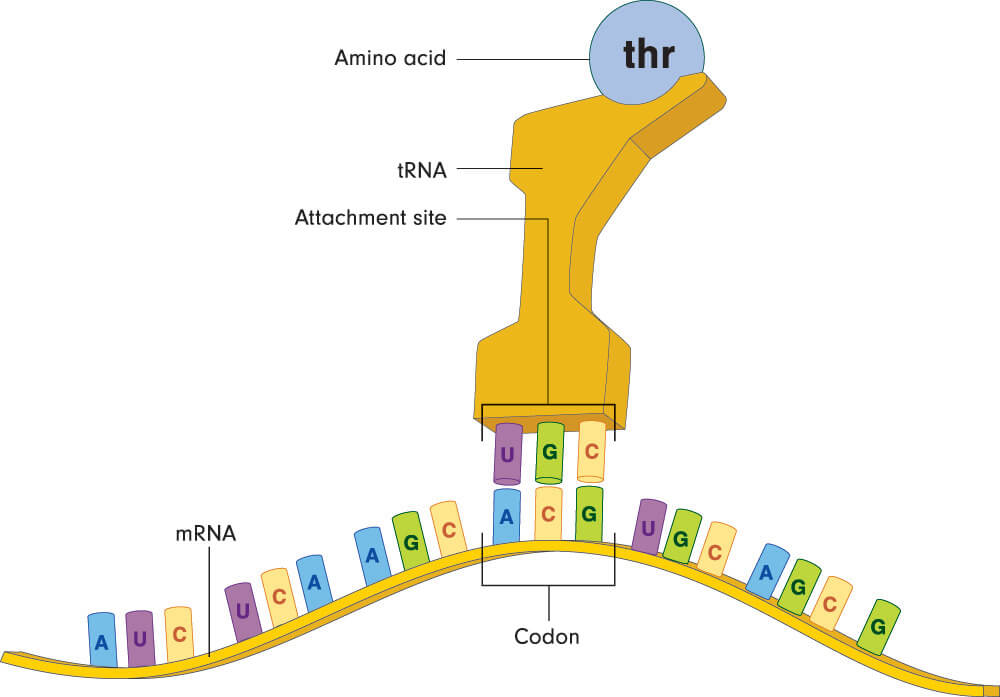
Each nucleotide triplet matches a single amino acid but most amino acids (except for methionine and tryptophan) match multiple codons. Leucine, serine, and arginine have six different codes per amino acid. This makes it less likely to produce the wrong protein in the presence of a genetic mutation. As the strand of mRNA makes its way through the ribosome, each amino acid links to the next to form a polypeptide chain (protein). The sequence of amino acids determines the type of protein, just as the sequence of nucleotide bases determines the type and order of the amino acids.
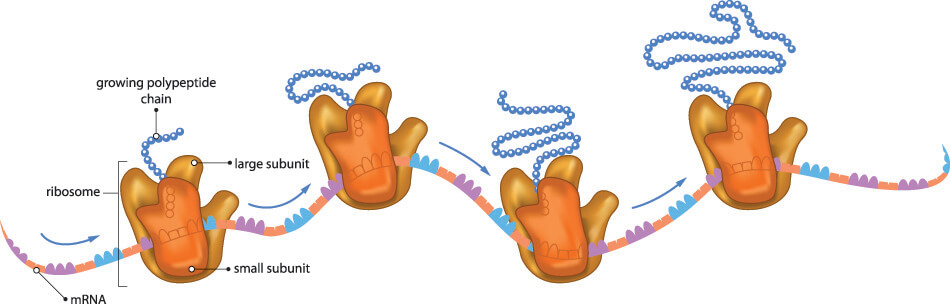
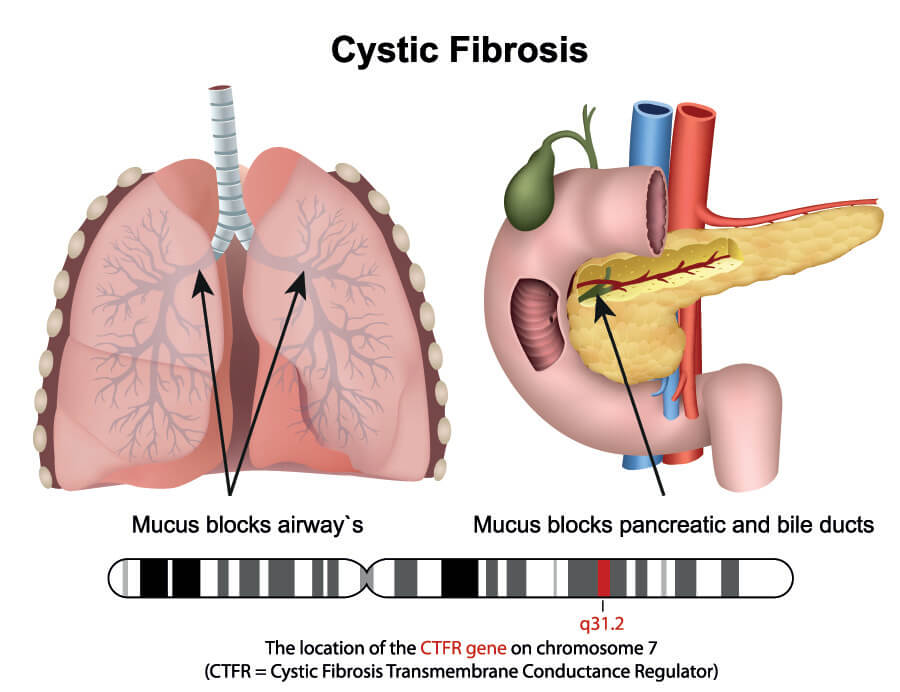
If no stop codons are present, a polypeptide chain continues to grow until there are no codons left on the mRNA strand. DNA transcription is not an exact process and the invisible stop codon in DNA (invisible because it is not understood or read) is followed by a long tail of nucleotide bases. The absence of a stop codon during the translation phase of protein synthesis would mean that these excess bases, if in groups of three or more, match to amino acids and so change the form of the polypeptide chain, the type of protein produced, and that protein’s function. For example, a missing or faulty stop codon would change the instructions for type I keratin 9 production and code for a different (and bigger) protein. Alternatively, premature termination of a gene sequence due to stop codon-forming changes during DNA repair and transcription might create shorter or truncated proteins. Many diseases are caused by premature stop codons that change the physiology and/or anatomy of an organism; in humans, one of the many genetic causes of cystic fibrosis is a stop codon mutation.
The detection of a stop codon signals the end of the translation process. The polypeptide chain is released from the ribosome and sent to where it can be used, either inside the cell or outside.
What are the Three Stop Codons?
The three stop codons are TAG, TAA, and TGA in sense DNA, and UAG, UAA, and UGA in mRNA.
TAG and UAG are called amber stop codons; TAA and UAA are known as ochre stop codons, and TGA and UGA are given the name of opal stop codons (or umber stop codons). The amber color code is attributed to the name of the scientist who first discovered it; the other colors simply continue this color-coded theme. Stop codons are also called nonsense codons or termination codons, the first of these terms because stop codons never code for amino acids, and the second due to stop codon function.
Stop Codon Mutations
Stop codon mutations can easily occur, especially when we consider the length of the genome and the thousands of different nucleotide triplets. Both transcription and translation processes are susceptible to a broad range of potential errors that may or may not lead to anatomical and physiological changes. The insertion of the wrong nucleotide in the KRT-9 gene in family members already predisposed to the disease has been found to contribute to the development of a skin disease known as epidermolytic palmoplantar keratoderma.

Which type of mutation creates a stop codon? Radiation, chemicals, pollution, infection, and the aging process are just some ways in which the DNA can become damaged; attempts to repair this damage can accidentally insert the wrong nucleotide. This might change a triplet that would normally have coded for an amino acid into a stop codon. When this happens, the result is a nonsense mutation. A nonsense mutation specifically changes an amino acid-producing triplet into a stop codon and leads to premature termination of protein synthesis in the ribosome.
While all kinds of mutations occur during DNA to mRNA transcription, mRNA only copies what is written without ever needing to understand it. For the period where mRNA is not in contact with a ribosome, even multiple mutations will not cause an effect. Effects are only seen when the changed code is translated into a faulty protein This is why most mutations are labeled as being part of the translation process, where the edited code may or may not produce a different amino acid. The fact that most amino acids match up to six different nucleotide triplets means there is a chance that, even in the presence of a mutation, the same protein will be produced. We usually associate genetic mutations with illness; however, they are also responsible for successful evolution. Genetic mutations help organisms to adapt to their environment.
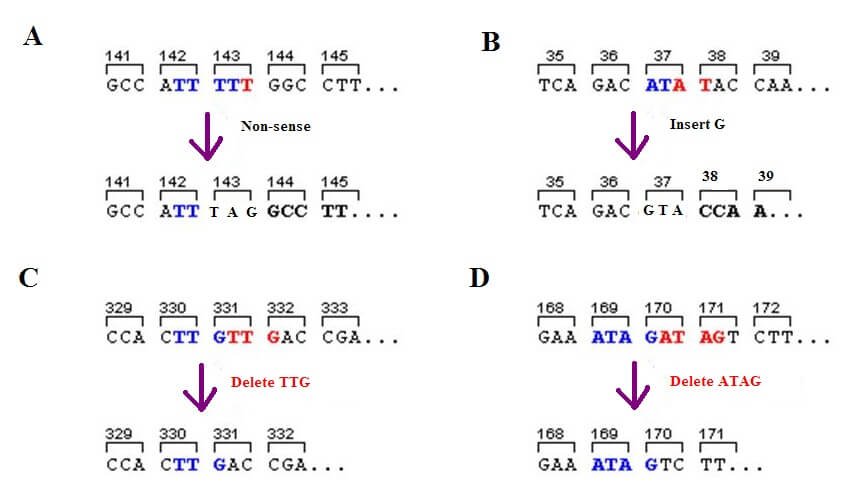
There are various forms of genetic mutation. Deletion mutations do not copy certain parts of the genome and so change the order of the nucleotides. A single base or multiple bases may be completely missed out. Insertion mutations add one or more nucleotides and also change the order of the genetic code. Substitution mutations (silent, missense, and nonsense) swap a single nucleotide (not multiple nucleotides) with a different base and this may or may not substitute a different amino acid in a polypeptide chain. If the same protein is produced, even in the presence of a mutation, it is called a silent mutation. In some cases, an entire section of DNA can swap between the two strands – this is called translocation.
If a different amino acid is added to the polypeptide chain that may or may not change its function, the cause is a missense mutation. Where substitution creates a stop codon by changing the code of a nucleotide triplet that matches an amino acid, it is called a nonsense mutation. The below image shows three types of mutation: A is the nonsense mutation, B the insertion mutation, and C and D show deletion mutations.
Quiz