Tetrameric protein

A tetrameric protein is a protein with a quaternary structure of four subunits (tetrameric). Homotetramers have four identical subunits (such as glutathione S-transferase), and heterotetramers are complexes of different subunits. A tetramer can be assembled as dimer of dimers with two homodimer subunits (such as sorbitol dehydrogenase), or two heterodimer subunits (such as hemoglobin).
Subunit interactions in tetramers
[edit]The interactions between subunits forming a tetramer is primarily determined by non covalent interaction.[1] Hydrophobic effects, hydrogen bonds and electrostatic interactions are the primary sources for this binding process between subunits. For homotetrameric proteins such as sorbitol dehydrogenase (SDH), the structure is believed to have evolved going from a monomeric to a dimeric and finally a tetrameric structure in evolution. The binding process in SDH and many other tetrameric enzymes can be described by the gain in free energy which can be determined from the rate of association and dissociation.[1] The above image shows the assembly of the four subunits (A,B,C and D) in SDH.
Hydrogen bonds between subunits
[edit]Hydrogen bonding networks between subunits has been shown to be important for the stability of the tetrameric quaternary protein structure. For example, a study of SDH which used diverse methods such as protein sequence alignments, structural comparisons, energy calculations, gel filtration experiments and enzyme kinetics experiments, could reveal an important hydrogen bonding network which stabilizes the tetrameric quaternary structure in mammalian SDH.[1]
Tetramers in immunology
[edit]In immunology, MHC tetramers can be used in tetramer assays, to quantify numbers of antigen-specific T cells (especially CD8+ T cells). MHC tetramers are based on recombinant class I molecules that, through the action of bacterial BirA, have been biotinylated. These molecules are folded with the peptide of interest and β2M and tetramerized by a fluorescently labeled streptavidin. (Streptavidin binds to four biotins per molecule.) This tetramer reagent will specifically label T cells that express T cell receptors that are specific for a given peptide-MHC complex. For example, a Kb/FAPGNYPAL tetramer will specifically bind to Sendai virus specific cytotoxic T cell in a C57BL/6 mouse. Antigen specific responses can be measured as CD8+, tetramer+ T cells as a fraction of all CD8+ lymphocytes.
The reason for using a tetramer, as opposed to a single labeled MHC class I molecule is that the tetrahedral tetramers can bind to three TCRs at once, allowing specific binding in spite of the low (1 micromolar) affinity of the typical class I-peptide-TCR interaction. MHC class II tetramers can also be made, although these are more difficult to work with practically.[2]
Homotetramers and heterotetramers
[edit]
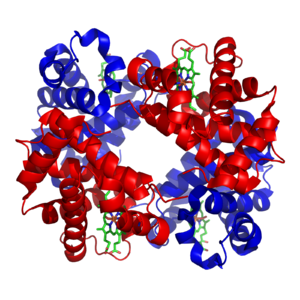
A homotetramer is a protein complex made up of four identical subunits which are associated but not covalently bound.[3] Conversely, a heterotetramer is a 4-subunit complex where one or more subunits differ.[4]
Examples of homotetramers include:
- enzymes like beta-glucuronidase (pictured)
- export factors such as SecB from Escherichia coli[5]
- magnesium ion transporters such as CorA.[6]
- lectins such as Concanavalin A
- IMPDH and IMPDH2
Examples of heterotetramers include haemoglobin (pictured), the NMDA receptor, some aquaporins,[7] some AMPA receptors, as well as some enzymes.[8]
Purification of heterotetramers
[edit]Ion-exchange chromatography is useful for isolating specific heterotetrameric protein assemblies, allowing purification of specific complexes according to both the number and the position of charged peptide tags.[9][10] Nickel affinity chromatography may also be employed for heterotetramer purification.[11]
Intragenic complementation
[edit]Multiple copies of a polypeptide encoded by a gene often can form an aggregate referred to as a multimer. When a multimer is formed from polypeptides produced by two different mutant alleles of a particular gene, the mixed multimer may exhibit greater functional activity than the unmixed multimers formed by each of the mutants alone. When a mixed multimer displays increased functionality relative to the unmixed multimers, the phenomenon is referred to as intragenic complementation. In humans, argininosuccinate lyase (ASL) is a homotetrameric enzyme that can undergo intragenic complementation. An ASL disorder in humans can arise from mutations in the ASL gene, particularly mutations that affect the active site of the tetrameric enzyme. ASL disorder is associated with considerable clinical and genetic heterogeneity which is considered to reflect the extensive intragenic complementation occurring among different individual patients.[12][13][14]
References
[edit]- ^ a b c Hellgren M, Kaiser C, de Haij S, Norberg A, Höög JO (December 2007). "A hydrogen-bonding network in mammalian sorbitol dehydrogenase stabilizes the tetrameric state and is essential for the catalytic power". Cellular and Molecular Life Sciences. 64 (23): 3129–3138. doi:10.1007/s00018-007-7318-1. PMC 11136444. PMID 17952367. S2CID 22090973.
- ^ Dolton G, Tungatt K, Lloyd A, Bianchi V, Theaker SM, Trimby A, et al. (September 2015). "More tricks with tetramers: a practical guide to staining T cells with peptide-MHC multimers". Immunology. 146 (1): 11–22. doi:10.1111/imm.12499. PMC 4552497. PMID 26076649.
- ^ "GO term: protein homotetramerization". YeastGenome. Archived from the original on 27 September 2011. Retrieved 14 May 2011.
- ^ "GO term: protein heterotetramerization". YeastGenome. Archived from the original on 27 September 2011. Retrieved 14 May 2011.
- ^ Watanabe M, Blobel G (April 1989). "Cytosolic factor purified from Escherichia coli is necessary and sufficient for the export of a preprotein and is a homotetramer of SecB". Proceedings of the National Academy of Sciences of the United States of America. 86 (8): 2728–2732. Bibcode:1989PNAS...86.2728W. doi:10.1073/pnas.86.8.2728. PMC 286991. PMID 2649892.
- ^ Warren MA, Kucharski LM, Veenstra A, Shi L, Grulich PF, Maguire ME (July 2004). "The CorA Mg2+ transporter is a homotetramer". Journal of Bacteriology. 186 (14): 4605–4612. doi:10.1128/JB.186.14.4605-4612.2004. PMC 438605. PMID 15231793.
- ^ Neely JD, Christensen BM, Nielsen S, Agre P (August 1999). "Heterotetrameric composition of aquaporin-4 water channels". Biochemistry. 38 (34): 11156–11163. doi:10.1021/bi990941s. PMID 10460172.
- ^ Chang TH, Hsieh FL, Ko TP, Teng KH, Liang PH, Wang AH (February 2010). "Structure of a heterotetrameric geranyl pyrophosphate synthase from mint (Mentha piperita) reveals intersubunit regulation". The Plant Cell. 22 (2): 454–467. doi:10.1105/tpc.109.071738. PMC 2845413. PMID 20139160.
- ^ Sakash JB, Kantrowitz ER (September 2000). "The contribution of individual interchain interactions to the stabilization of the T and R states of Escherichia coli aspartate transcarbamoylase". The Journal of Biological Chemistry. 275 (37): 28701–28707. doi:10.1074/jbc.M005079200. PMID 10875936.
- ^ Fairhead M, Krndija D, Lowe ED, Howarth M (January 2014). "Plug-and-play pairing via defined divalent streptavidins". Journal of Molecular Biology. 426 (1): 199–214. doi:10.1016/j.jmb.2013.09.016. PMC 4047826. PMID 24056174.
- ^ Howarth M, Chinnapen DJ, Gerrow K, Dorrestein PC, Grandy MR, Kelleher NL, et al. (April 2006). "A monovalent streptavidin with a single femtomolar biotin binding site". Nature Methods. 3 (4): 267–273. doi:10.1038/nmeth861. PMC 2576293. PMID 16554831.
- ^ Turner MA, Simpson A, McInnes RR, Howell PL (August 1997). "Human argininosuccinate lyase: a structural basis for intragenic complementation". Proceedings of the National Academy of Sciences of the United States of America. 94 (17): 9063–9068. Bibcode:1997PNAS...94.9063T. doi:10.1073/pnas.94.17.9063. PMC 23030. PMID 9256435.
- ^ Yu B, Howell PL (October 2000). "Intragenic complementation and the structure and function of argininosuccinate lyase". Cellular and Molecular Life Sciences. 57 (11): 1637–1651. doi:10.1007/PL00000646. PMC 11147086. PMID 11092456. S2CID 1254964.
- ^ Yu B, Thompson GD, Yip P, Howell PL, Davidson AR (December 2001). "Mechanisms for intragenic complementation at the human argininosuccinate lyase locus". Biochemistry. 40 (51): 15581–15590. doi:10.1021/bi011526e. PMID 11747433.