Protein engineering
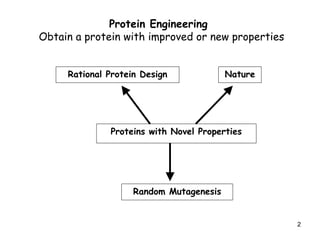
- 2. 2 Protein Engineering Obtain a protein with improved or new properties Rational Protein Design Nature Proteins with Novel Properties Random Mutagenesis
- 3. 3 Protein Engineering -> Mutagenesis used for modifying proteins Replacements on protein level -> mutations on DNA level Assumption : Natural sequence can be modified to improve a certain function of protein This implies: • Protein is NOT at an optimum for that function • Sequence changes without disruption of the structure • (otherwise it would not fold) • New sequence is not TOO different from the native sequence (otherwise loss in function of protein) consequence -> introduce point mutations
- 4. 4 Mutagenesis Mutagenesis -> change in DNA sequence -> Point mutations or large modifications Point mutations (directed mutagenesis): - Substitution: change of one nucleotide (i.e. A-> C) - Insertion: gaining one additional nucleotide - Deletion: loss of one nucleotide
- 5. Consequences of point mutations within a coding 5 sequence (gene) for the protein Silent mutations: -> change in nucleotide sequence with no consequences for protein sequence -> Change of amino acid -> truncation of protein -> change of c-terminal part of protein -> change of c-terminal part of protein
- 6. 6 Applications of directed mutagenesis
- 7. 7 Approaches for directed mutagenesis -> site-directed mutagenesis -> point mutations in particular known area result -> library of wild-type and mutated DNA (site-specific) not really a library -> just 2 species -> random mutagenesis -> point mutations in all areas within DNA of interest result -> library of wild-type and mutated DNA (random) a real library -> many variants -> screening !!! if methods efficient -> mostly mutated DNA
- 8. 8 Rational Protein Design Þ Site –directed mutagenesis !!! Requirements: -> Knowledge of sequence and preferable Structure (active site,….) -> Understanding of mechanism (knowledge about structure – function relationship) -> Identification of cofactors……..
- 9. 9 Site-directed mutagenesis methods
- 10. 10 Site-directed mutagenesis methods – Oligonucleotide - directed method
- 11. Site-directed mutagenesis methods – PCR based 11
- 12. Directed Evolution – Random mutagenesis 12 -> based on the process of natural evolution - NO structural information required - NO understanding of the mechanism required General Procedure: Generation of genetic diversity Þ Random mutagenesis Identification of successful variants Þ Screening and seletion
- 13. 13
- 14. 14 Random Mutagenesis (PCR based) with degenerated primers (saturation mutagenesis)
- 15. 15 Random Mutagenesis (PCR based) with degenerated primers (saturation mutagenesis)
- 16. 16 Random Mutagenesis (PCR based) Error –prone PCR -> PCR with low fidelity !!! Achieved by: - Increased Mg2+ concentration - Addition of Mn2+ - Not equal concentration of the four dNTPs - Use of dITP - Increasing amount of Taq polymerase (Polymerase with NO proof reading function)
- 17. 17 Random Mutagenesis (PCR based) DNA Shuffling DNase I treatment (Fragmentation, 10-50 bp, Mn2+) Reassembly (PCR without primers, Extension and Recombination) PCR amplification
- 18. 18 Random Mutagenesis (PCR based) Family Shuffling Genes coming from the same gene family -> highly homologous -> Family shuffling
- 19. 19 Protein Engineering What can be engineered in Proteins ? -> Folding (+Structure): 1. Thermodynamic Stability (Equilibrium between: Native Û Unfolded state) 2. Thermal and Environmental Stability (Temperature, pH, Solvent, Detergents, Salt …..)
- 20. 20 Protein Engineering What can be engineered in Proteins ? -> Function: 1. Binding (Interaction of a protein with its surroundings) How many points are required to bind a molecule with high affinity? 2. Catalysis (a different form of binding – binding the transition state of a chemical reaction) Increased binding to the transition state Þ increased catalytic rates !!! Requires: Knowledge of the Catalytic Mechanism !!! -> engineer Kcat and Km
- 21. 21 Protein Engineering Factors which contribute to stability: 1. Hydrophobicity (hydrophobic core) 2. Electrostatic Interactions: -> Salt Bridges -> Hydrogen Bonds -> Dipole Interactions 3. Disulfide Bridges 4. Metal Binding (Metal chelating site) 5. Reduction of the unfolded state entropy with X ® Pro mutations
- 22. 22 Protein Engineering Design of Thermal and Environmental stability: 1. Stabilization of a-Helix Macrodipoles 2. Engineer Structural Motifes (like Helix N-Caps) 3. Introduction of salt bridges 4. Introduction of residues with higher intrinsic properties for their conformational state (e.g. Ala replacement within a a-Helix) 5. Introduction of disulfide bridges 6. Reduction of the unfolded state entropy with X ® Pro mutations
- 23. 23 Protein Engineering - Applications Engineering Stability of Enzymes – T4 lysozyme -> S-S bonds introduction
- 24. 24 Protein Engineering - Applications Engineering Stability of Enzymes – triosephosphate isomerase from yeast -> replace Asn (deaminated at high temperature)
- 25. 25 Protein Engineering - Applications Site-directed mutagenesis -> used to alter a single property Problem : changing one property -> disrupts another characteristics Directed Evolution (Molecular breeding) -> alteration of multiple properties
- 26. 26 Protein Engineering – Applications Directed Evolution
- 27. 27 Protein Engineering – Applications Directed Evolution
- 28. 28 Protein Engineering – Directed Evolution
- 29. 29 Protein Engineering - Applications