The New York Genome Center’s ALS Consortium
Our ALS Consortium – a partnership of clinicians, basic scientists, geneticists, and computational biologists from 45 institutions across the globe – establishes a framework to apply whole genome sequencing and functional genomics to the study of ALS.
Global Collaboration to Find a Cure
ALS-causing mutations tend to be of moderate impact and are relatively rare in the population, and identification of new mutations requires several thousand samples. As even the busiest ALS clinics see only 400 patients per year, samples need to be obtained from multiple sites. The ALS consortium facilitates this collaboration between sites to enable broad data sharing. This joint effort has sequenced more than 4,359 whole genomes and 2,348 RNA samples. These data, and data mining tools, are shared both within our consortium and with other consortia, with the aim of finding novel ALS-associated mutations. These efforts have contributed to many published studies, including the identification of novel ALS-associated mutations such as mutations in the KIF5A gene.
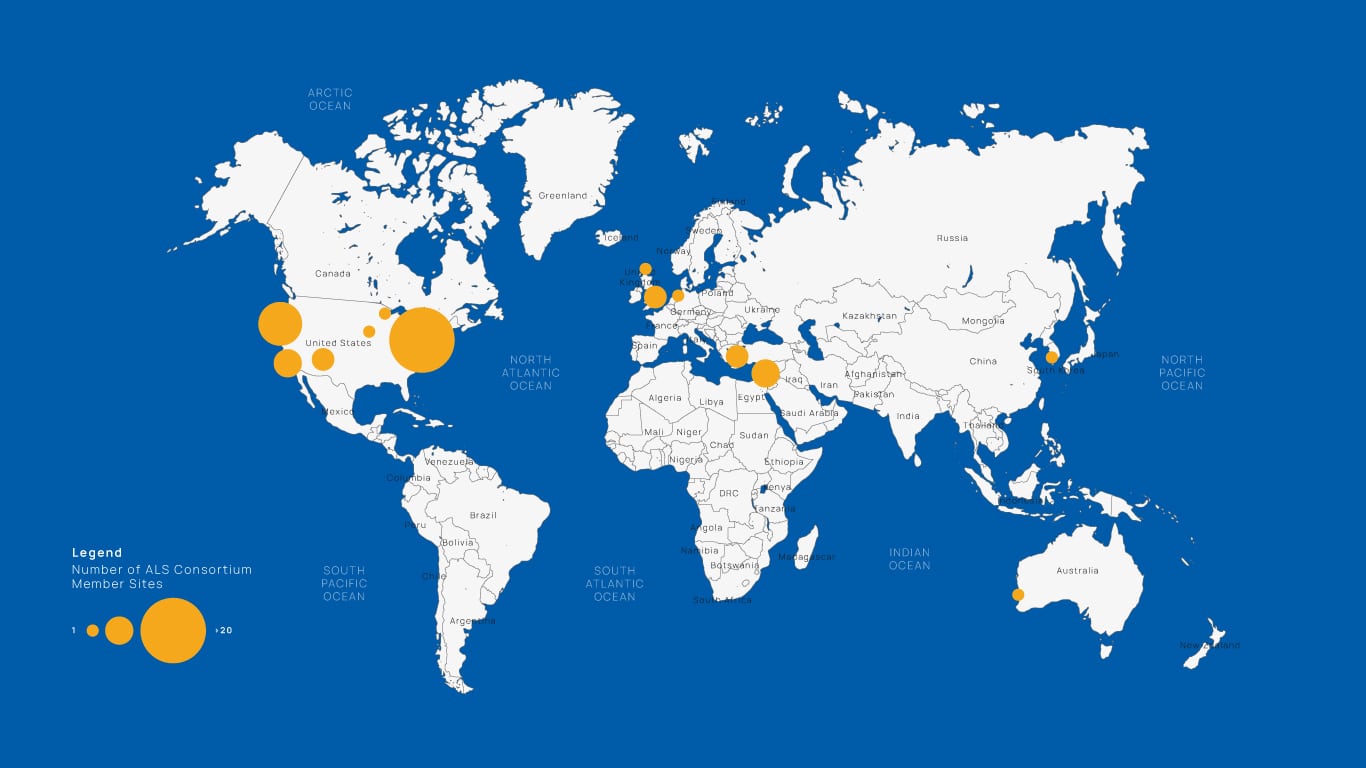
ALS Consortium Member Sites
NYGC ALS Consortium Goals
Broadly stated, the goals of this consortium are to integrate whole genome sequencing with RNA sequencing so as to interrogate relationships between mutations, gene expression, and disease mechanisms. The consortium will further integrate genomic data with clinical data on postmortem tissue and histopathology so as to be able to study the molecular mechanisms causing the disease to spread.
Such work will eventually help identify mutations associated with different forms of the disease, or gene variants that can modify the presentation of the disease and could be further studied to identify pathways for the targeted development of therapies. Finally, the consortium hopes to create and maintain a data warehouse for genomic data that can be broadly accessed by the academic community.
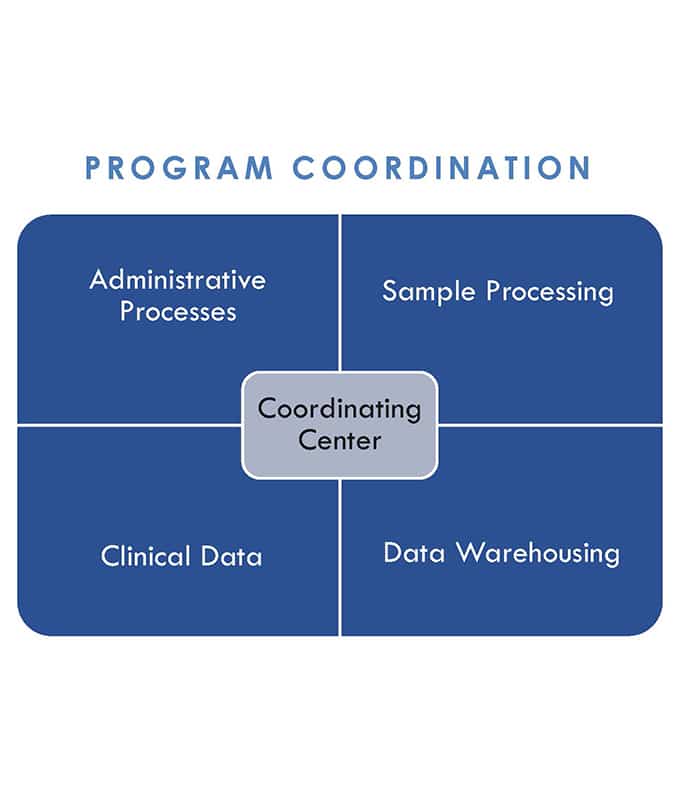
ALS Consortium Coordination
To enable broad data sharing and smooth coordination between sites, we centralize the administration and operation of a widespread, highly federated program. We disseminate key elements for Informed Consent Forms and IRB protocols, and common data elements to harmonize clinical phenotyping across participating centers. Additionally, data and novel analytical data mining tools are shared both within our consortium and with other ALS consortia. These collaborative efforts are geared towards making the best use of sequencing data to ultimately facilitate a more efficient pursuit of novel ALS treatments. Comparing clinical profiles to genomic profiles can enable us to determine whether specific mutations are associated with specific clinical outcomes to enable patient-specific medicine in the future.
ALS Consortium Members to Date
- New York Genome Center
- The University of Maryland
- Cedars-Sinai Medical Center
- Pennsylvania State University
- Henry Ford Health System
- University of Pennsylvania
- Columbia University
- Massachusetts Institute of Technology
- Johns Hopkins University
- Academic Medical Center, University of Amsterdam
- University of California, San Francisco
- Massachusetts General Hospital
- University College London / Queen Mary University of London
- The Jackson Laboratory
- University of California, Irvine
- Gladstone Institutes
- University of Thessaly
- Washington University in St. Louis
- The University of Edinburgh
- Weizmann Institute of Science
- Icahn School of Medicine at Mount Sinai
- Temple University
- Cold Spring Harbor Laboratory
- Brigham and Women’s Hospital
- The Research Foundation for the State University of New York (Stony Brook)
- NIH / National Institute of Neurological Disorders and Strokes
- Hospital for Special Surgery
- Barrow Neurological Institute
- Regeneron
- Atlantic Health System
- University of Athens
- Hadassah Hebrew University
- Georgetown University
- Tel Aviv Sourasky Medical Center
- University of California, San Diego
- The Translational Genomics Research Institute (TGen)
- Broad Institute
- Norwell Health
- Korea Advanced Institute of Science and Technology
- Maze Therapeutics
- Bristol Myers Squibb
- NIH / National Institute of Allergy and Infectious Disease
- Perron Institute for Neurological and Translational Science
- Pfizer
Latest News & Publications
-
Nature Medicine · September 3, 2024
-
Muscle & Nerve · August 9, 2024
Mitochondrial genome variants associated with amyotrophic lateral sclerosis and their haplogroup distribution.
-
bioRxiv · July 19, 2024 · Preprint
ALS molecular subtypes are a combination of cellular, genetic, and pathological features learned by deep multiomics classifiers.
Acknowledgements
The patients who participated in these studies.
Contact Us

