Serine protease
- 1. Serine Proteases Gisha G P S 3 MSc Biotechnology
- 3. Classification of proteases • Serine proteases • Cystein proteases • Aspartate proteases • Metallo proteases
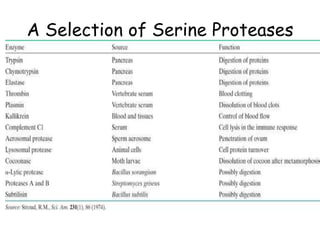
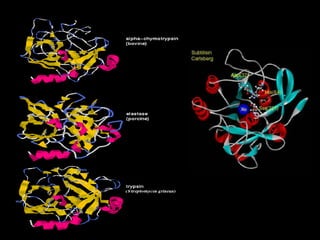
- 4. A Selection of Serine Proteases
- 5. Introduction • Large family of proteolytic enzymes • All have serine residue at their active site which plays a crucial part in the enzymatic activity. • All cleave peptide bonds, by a similar mechanism of action. They differ in their specificity and regulation.
- 6. Serine proteases include: • the pancreatic proteases: trypsin, chymotrypsin and elastase, • various tissue/intracellular proteases such as leukocyte elastase • enzymes of the blood clotting cascade • some enzymes of complement system • Many serine proteases are synthesized as inactive precursors (zymogens) which are activated by proteolysis
- 7. Classification • Trypsin like serine peptidase – if it prefers to cleave peptide bonds of lys & arg • Chymotrypsin like serine peptidase – if it prefers to cleave aromatic amino acids • Elastase like serine peptidase - if it prefers to cleave amino acids with small side chain groups like ala
- 8. • Subtilisin like serine peptidase – serine protease in prokaryotes. • Very different primary and tertiary structures fro those of the mammalian proteases. • The active site structures and mechanism of action of all these enzymes are almost identical.
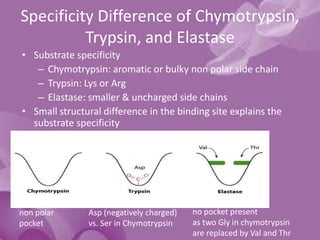
- 10. Specificity Difference of Chymotrypsin, Trypsin, and Elastase • Substrate specificity – Chymotrypsin: aromatic or bulky non polar side chain – Trypsin: Lys or Arg – Elastase: smaller & uncharged side chains • Small structural difference in the binding site explains the substrate specificity non polar pocket Asp (negatively charged) vs. Ser in Chymotrypsin no pocket present as two Gly in chymotrypsin are replaced by Val and Thr
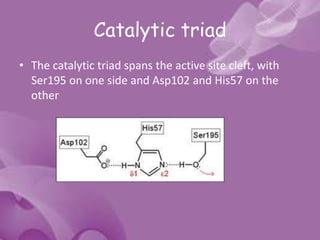
- 12. The Catalytic Components 1. The Catalytic Triad 2. The Oxyanion Hole
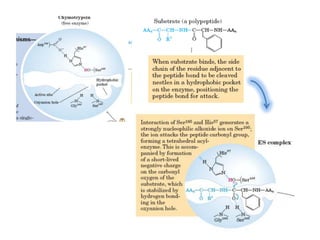
- 13. Catalytic triad • The catalytic triad spans the active site cleft, with Ser195 on one side and Asp102 and His57 on the other
- 14. • The catalytic triad is part of an extensive hydrogen bonding network • Hydrogen bonds are generally observed between the Nδ1-H of His57 and Oδ1 of Asp102 and between the OH of Ser195 and the Nε2-H of His57.
- 15. The Oxyanion Hole • The oxyanion hole is formed by the backbone NHs of Gly193 and Ser195.
- 16. • These atoms form a pocket of positive charge that activates the carbonyl of the scissile peptide bond and stabilizes the negatively charged oxyanion of the tetrahedral intermediate • Engages the backbone O atom of the P1 residue of substrate in an important H- bonding interaction.
- 17. The Substrate Recognition Sites • The substrate recognition sites include the polypeptide binding site and the binding pockets for the side chains of the peptide substrate • The active site of serine proteases is shaped as a cleft where the polypeptide substrate binds.
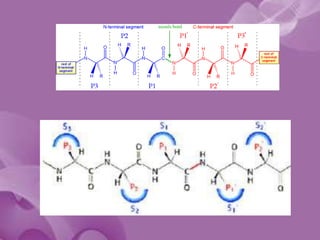
- 18. • Schechter and Berger [1] labeled amino acid residues from N to C term of the polypeptide substrate (Pi, ..., P3, P2, P1, P1', P2', P3', ..., Pj) and their respective binding sub-sites Si,..., S3, S2, S1, S1', S2', S3',..., Sj) . The cleavage is catalyzed between P1 and P1'.
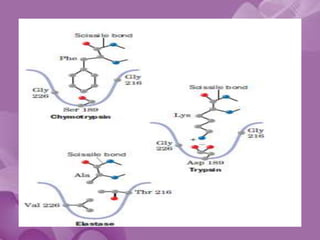
- 20. The S1 Site • Specificity of chymotrypsin-like serine proteases is usually categorized in terms of the P1-S1 interaction. • The S1 site is a pocket adjacent to Ser195, formed by residues 189-192, 214-216, and 224-228 • Specificity is usually determined by the residues at positions 189, 216, and 226
- 21. • The combination of Ser189, Gly216, and Gly226 create a deep hydrophobic pocket in chymotrypsin that accounts for this specificity. • Asp189, Gly216, and Gly226 create a negatively charged S1 site that accounts for trypsin’s specificity for substrates containing Arg or Lys at P1. • Elastase prefers substrates with small aliphatic residues at P1; the S1 site of elastase is smaller than the S1 sites of chymotrypsin and trypsin due to the presence of Val216 and Thr226.
- 22. The Polypeptide Binding Site • The polypeptide binding site refers to the main chain of residues 214-216 which form an anti parallel beta sheet with the backbone of the P1-P3 residues of a peptide substrate • In chymotrypsin, hydrogen bonds form between the carbonyl oxygen of Ser214 and the NH of the P1 residue, the NH of Trp215 and the carbonyl of P3 and the carbonyl of Gly216 and the NH of P3. • These interactions are a general feature of chymotrypsin-like proteases and are critical for efficient substrate hydrolysis.
- 23. • Gly216 has different conformations in chymotrypsin, trypsin, and elastase, which suggests that the strength of this hydrogen bond will vary • Residues 214-216 also form one wall of the S1 site, and that the carbonyl of Ser214 forms a hydrogen bond to His57. • These structural interactions form a line of communication between the polypeptide binding site, the S1 site, and the catalytic triad.
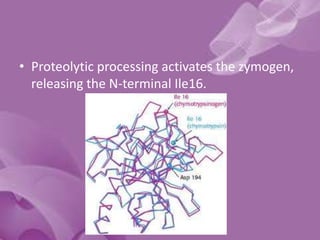
- 24. The Zymogen Activation Domain • Chymotrypsin-like proteases are synthesized as inactive precursors (“zymogens”) containing N-terminal extensions • Four segments are deformed in the zymogens of chymotrypsin and trypsin: the N-terminus to residue 19, residues 142-152, 184-193, and 216- 223 (these regions are collectively termed the activation domain36). • This deformed region includes the S1 site and oxyanion hole, which explains the low activity of the zymogen.
- 25. • Proteolytic processing activates the zymogen, releasing the N-terminal Ile16.
- 26. • The new N-terminus forms a buried salt bridge with Asp194, inducing a conformational change that orders the activation domain. • The S1 site and oxyanion hole are formed, creating the active protease.
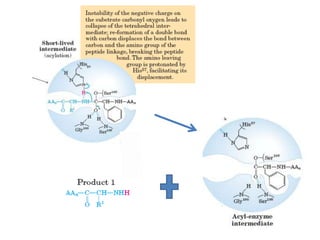
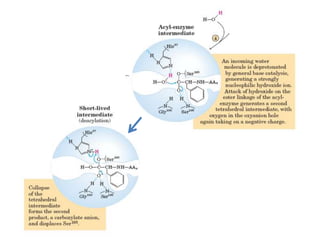
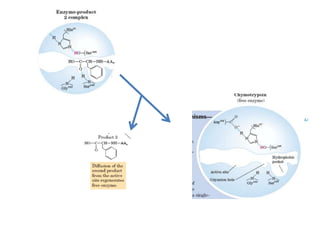
- 28. • The catalytic action of serine peptidases depends on the interplay of a nucleophile, a general base and an acid • Two steps: Acylation & Deacylation
- 30. =
- 33. References • Lehninger Principles of Biochemistry, 4E, David L. Nelson , Michael M. Cox • Fundamentals of Biochemistry, Voet & Voet • Serine Protease Mechanism and Specificity Lizbeth Hedstrom Department of Biochemistry, MS 009, Brandeis University, Waltham, Massachusetts 02454 • Functional role of catalytic triad and oxyanion hole-forming residues on enzyme activity of Escherichia coli thioesterase I/protease I/ phospholipase L1 Li-Chiun LEE*, Ya-Lin LEE†1, Ruey- Jyh LEU‡ and Jei-Fu SHAW











![• Schechter and Berger [1] labeled amino acid
residues from N to C term of the polypeptide
substrate (Pi, ..., P3, P2, P1, P1', P2', P3', ..., Pj)
and their respective binding sub-sites
Si,..., S3, S2, S1, S1', S2', S3',..., Sj) . The
cleavage is catalyzed between P1 and P1'.](https://image.slidesharecdn.com/serineprotease-140407002013-phpapp02/85/Serine-protease-18-320.jpg)